Samantha J. Gleich
Data Visualization
Amplicon Data (16S/18S)
Amplicon datasets are commonly used answer a key question in microbial ecology: "who is there?". These datasets can be visualized in a number of ways. Here, I will show you some of my favorite ways to visualize amplicon data in R and python.
Taxa Barplots - R
#install.packages('vegan')
#install.packages('ggplot2')
#install.packages('randomcoloR')
#install.packages('reshape2')
#install.packages('tidyverse')
library(vegan)
library(ggplot2)
library(randomcoloR)
library(reshape2)
library(tidyverse)
set.seed(10)
# Load data - Rows are samples and columns are features (species)
data(dune)
# Calculate relative abundances
dune$sum <- rowSums(dune)
dune <- dune/dune$sum
dune$sum <- NULL
rowSums(dune) # Check that row sums = 1
# Add a sample # column
dune$Sample <- 1:nrow(dune)
# Melt the dataframe
duneMelt <- melt(dune,id.vars=c("Sample"))
taxKey <- data.frame(variable=c(unique(duneMelt$variable)))
taxKey$Group <- as.factor(sample(5)) # Here, 'Group' denotes taxonomic groups (5 groups in this case).
# Add taxonomic information to the melted dataframe
duneMelt <- left_join(duneMelt,taxKey)
# Sum 'Group' relative abundance counts for each sample.
duneMeltSum <- duneMelt %>% group_by(Group,Sample) %>% summarize(s=sum(value)) %>% as.data.frame()
# Plot
ggplot(duneMeltSum,aes(x=Sample,y=s,fill=Group))+geom_bar(position="fill",stat="identity",color="grey20")+theme_classic()+xlab("Sample")+ylab("Relative Abundance")+scale_fill_manual(name="Taxonomic Group",values=c("red","blue","green","gold","grey"))+scale_x_continuous(breaks=seq(1:20),labels=seq(1:20))+theme(axis.text.x = element_text(angle=45,hjust =1))
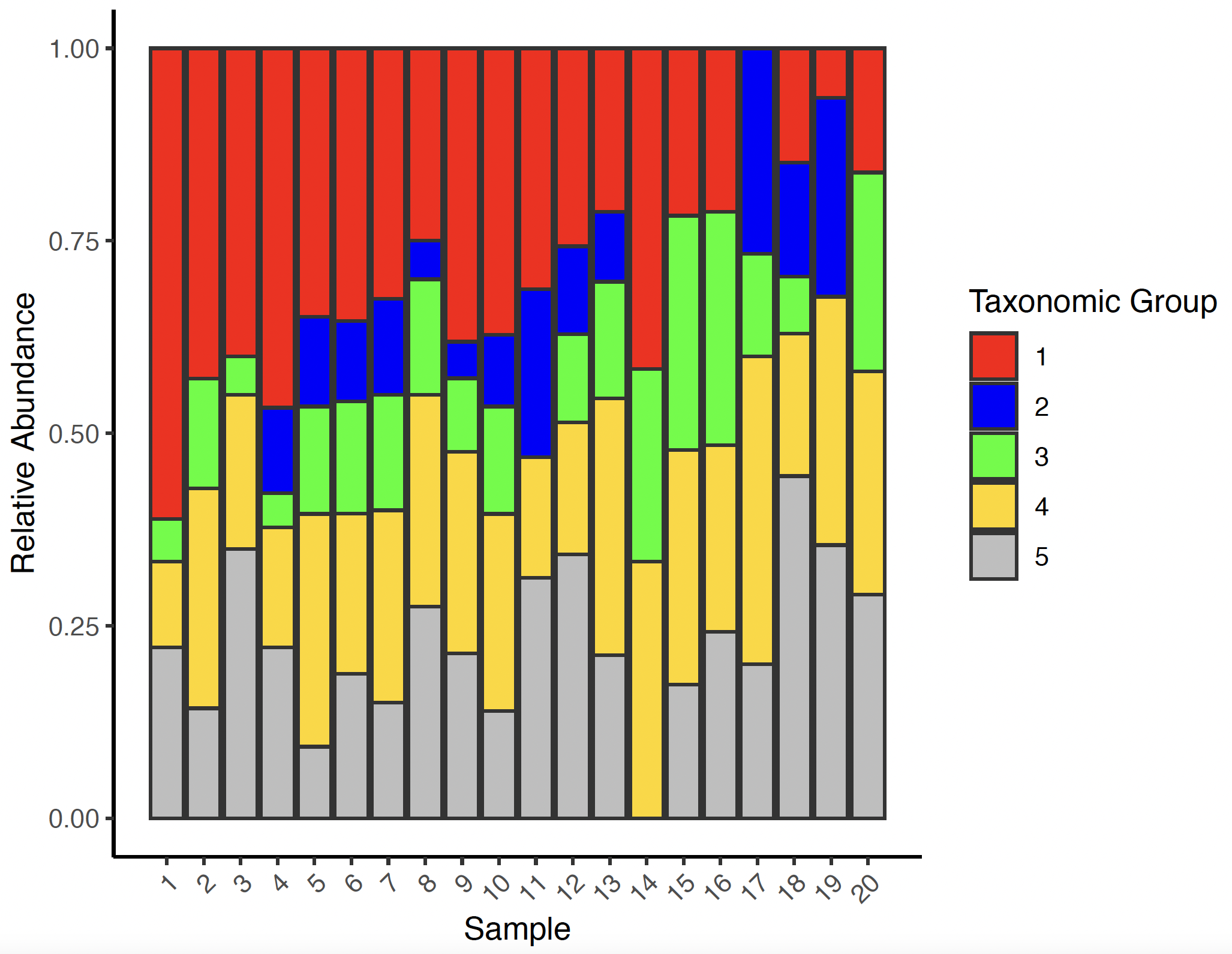
Taxa Barplots Over Time - R (Above Example Continued)
# Now let's assume that the sample numbers represent time points. We can remove some samples and use shading to show potential changes over time.
keep <- c(1,7,11,17,20)
duneMeltSum <- subset(duneMeltSum,Sample %in% keep)
# Plot
ggplot(duneMeltSum,aes(x=Sample,y=s,fill=Group))+geom_area(alpha = 0.4, position = 'fill')+geom_col(width = 1.5, color = 'gray20', position = 'fill')+scale_fill_manual(name="Taxonomic Group",values=c("red","blue","green","gold","grey"))+theme_classic()+xlab("Sample")+ylab("Relative Abundance")+theme(axis.text.x = element_text(angle = 45, hjust =1))+scale_x_continuous(breaks=c(unique(duneMeltSum$Sample)))

Taxa Barplots - Python
code.